NEW STUDY. A research group at the University of Gothenburg has developed a method that makes the exact binding sites of specific proteins in the genome discernible. The method is based on the idea that DNA damage induced by ultraviolet (UV) light can change where proteins bind.
The method is now published in the journal Nature Communications. Kerryn Elliott, a researcher in Erik Larsson Lekholm’s group at the Department for Medical Chemistry and Cell Biology at the Institute of Biomedicine, is the first author.
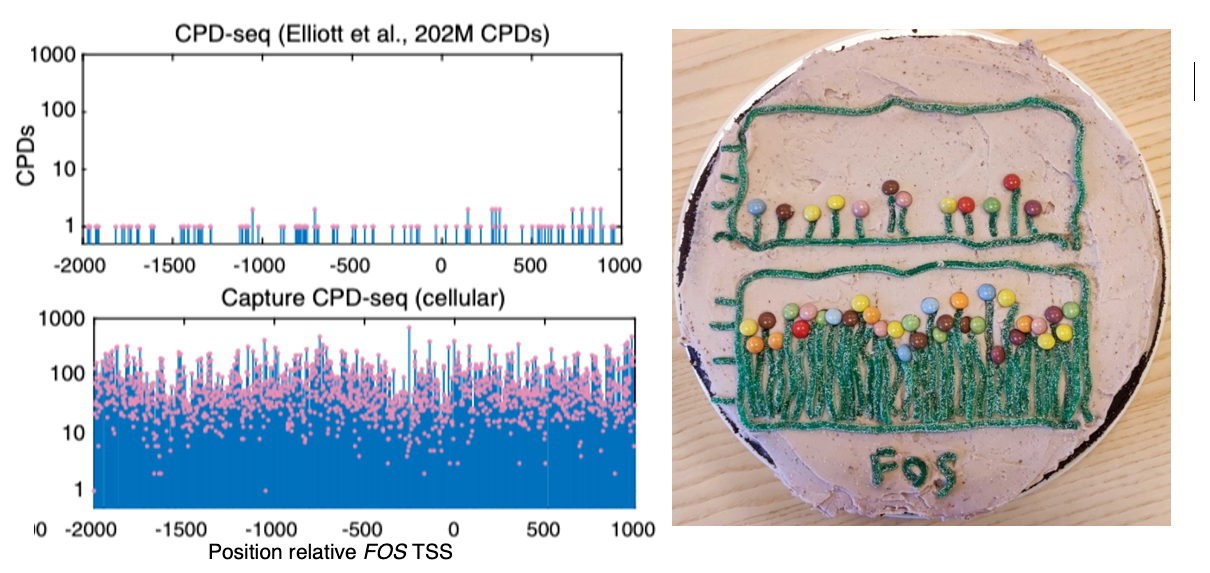
“The method allows us to map the footprints of several proteins simultaneously. It captures the interaction between protein and DNA at the moment the UV light reaches it,” Elliott says.
Clear binding profiles

The group developed the method in part to be able to study the origin of frequently occurring mutations in promoters (gene regulatory regions) in malignant melanoma. They found that UV-induced DNA damage, also known as cyclobutane pyrimidine dimers (CPDs), is strongly increased at certain protein binding sites. This phenomenon explains why mutations can arise repeatedly in promoters in skin cancers, and argues against such mutations being important in the development of cancer. The new method is the first time that UV damage has been precisely measured in DNA at the base level, and can also be used to map exactly where in the genome proteins bind.
“We get clear binding profiles for several DNA-binding proteins. We were even able to train a computational model that can predict whether a given sequence is bound or not based on its UV damage signature, which I think is pretty cool,” Elliott says.
Back-to-back publication
With the method, known as Capture CPD-seq, a previously established method called CPD-seq is combined with a “capture” of certain promoter sequences to allow for quantitative analysis of CPDs at individual DNA bases following next-generation sequencing at the Clinical Genomics Core facility. With the new method, the amount of UV damage in the DNA bound by proteins inside cells can be compared with protein-free DNA to create a “UV fingerprint”.
The hope is that the method can also be used to map various DNA structures and other interesting locations in the genome that have proved hard to analyze in cells.

The first people to perform the CPD-seq method with UV light was a research group at Washington State University that applied the method to studying UV damage in yeast. Erik Larsson Lekholm’s group has since adapted it for use in human cells. Last year, the two groups realized that they were both working on similar capture-based CPD-seq projects, and each of the groups has therefore written its own paper for Nature Communications. The two articles were published simultaneously.
“In the near future, we hope to use this approach to profile complete genomes. This may be possible for example in yeast, which has much smaller chromosomes than human cells.” Kerryn Elliott concludes.
Title: Base-resolution UV footprinting by sequencing reveals distinctive damage signatures for DNA-binding proteins; Nature Communications, https://doi.org/10.1038/s41467-023-38266-2.
BY: ELIN LINDSTRÖM